Bird’s eye view#
Background#
Data lineage tracks data’s journey, detailing its origins, transformations, and interactions to trace biological insights, verify experimental outcomes, meet regulatory standards, and increase the robustness of research. While tracking data lineage is easier when it is governed by deterministic pipelines, it becomes hard when its governed by interactive human-driven analyses.
Here, we’ll backtrace file transformations through notebooks, pipelines & app uploads in a research project based on Schmidt22 which conducted genome-wide CRISPR activation and interference screens in primary human T cells to identify gene networks controlling IL-2 and IFN-γ production.
Setup#
We need an instance:
!lamin init --storage ./mydata
Show code cell output
💡 creating schemas: core==0.46.1
✅ saved: User(id='DzTjkKse', handle='testuser1', email='testuser1@lamin.ai', name='Test User1', updated_at=2023-08-28 18:27:58)
✅ saved: Storage(id='lV7yGE5C', root='/home/runner/work/lamin-usecases/lamin-usecases/docs/mydata', type='local', updated_at=2023-08-28 18:27:58, created_by_id='DzTjkKse')
✅ loaded instance: testuser1/mydata
💡 did not register local instance on hub (if you want, call `lamin register`)
Import lamindb:
import lamindb as ln
✅ loaded instance: testuser1/mydata (lamindb 0.51.0)
We simulate the raw data processing of Schmidt22 with toy data in a real world setting with multiple collaborators (here testuser1 and testuser2):
assert ln.setup.settings.user.handle == "testuser1"
bfx_run_output = ln.dev.datasets.dir_scrnaseq_cellranger(
"perturbseq", basedir=ln.settings.storage, output_only=False
)
ln.track(ln.Transform(name="Chromium 10x upload", type="pipeline"))
ln.File(bfx_run_output.parent / "fastq/perturbseq_R1_001.fastq.gz").save()
ln.File(bfx_run_output.parent / "fastq/perturbseq_R2_001.fastq.gz").save()
Show code cell output
✅ saved: Transform(id='esPe9ey7UOWkUs', name='Chromium 10x upload', type='pipeline', updated_at=2023-08-28 18:27:59, created_by_id='DzTjkKse')
✅ saved: Run(id='vYYOHgAJkgeRJIuStm1D', run_at=2023-08-28 18:27:59, transform_id='esPe9ey7UOWkUs', created_by_id='DzTjkKse')
💡 file in storage 'mydata' with key 'fastq/perturbseq_R1_001.fastq.gz'
💡 file in storage 'mydata' with key 'fastq/perturbseq_R2_001.fastq.gz'
Track a bioinformatics pipeline#
When working with a pipeline, we’ll register it before running it.
This only happens once and could be done by anyone on your team.
ln.setup.login("testuser2")
✅ logged in with email testuser2@lamin.ai and id bKeW4T6E
❗ record with similar name exist! did you mean to load it?
| id | __ratio__ | |
|---|---|---|
| name | ||
| Test User1 | DzTjkKse | 90.0 |
✅ saved: User(id='bKeW4T6E', handle='testuser2', email='testuser2@lamin.ai', name='Test User2', updated_at=2023-08-28 18:28:00)
transform = ln.Transform(name="Cell Ranger", version="7.2.0", type="pipeline")
ln.User.filter().df()
| handle | name | updated_at | ||
|---|---|---|---|---|
| id | ||||
| DzTjkKse | testuser1 | testuser1@lamin.ai | Test User1 | 2023-08-28 18:27:58 |
| bKeW4T6E | testuser2 | testuser2@lamin.ai | Test User2 | 2023-08-28 18:28:00 |
transform
Transform(id='JXkdP1eEWX0T9R', name='Cell Ranger', version='7.2.0', type='pipeline', created_by_id='bKeW4T6E')
ln.track(transform)
✅ saved: Transform(id='JXkdP1eEWX0T9R', name='Cell Ranger', version='7.2.0', type='pipeline', updated_at=2023-08-28 18:28:00, created_by_id='bKeW4T6E')
✅ saved: Run(id='gXXVcK1otXqE6UHukFqd', run_at=2023-08-28 18:28:00, transform_id='JXkdP1eEWX0T9R', created_by_id='bKeW4T6E')
Now, let’s stage a few files from an instrument upload:
files = ln.File.filter(key__startswith="fastq/perturbseq").all()
filepaths = [file.stage() for file in files]
💡 adding file yyzQ5nQh9AADZSJHeO94 as input for run gXXVcK1otXqE6UHukFqd, adding parent transform esPe9ey7UOWkUs
💡 adding file T2qbXLg7u11pNkJ9aoWC as input for run gXXVcK1otXqE6UHukFqd, adding parent transform esPe9ey7UOWkUs
Assume we processed them and obtained 3 output files in a folder 'filtered_feature_bc_matrix':
output_files = ln.File.from_dir("./mydata/perturbseq/filtered_feature_bc_matrix/")
ln.save(output_files)
Show code cell output
✅ created 3 files from directory using storage /home/runner/work/lamin-usecases/lamin-usecases/docs/mydata and key = perturbseq/filtered_feature_bc_matrix/
Let’s look at the data lineage at this stage:
output_files[0].view_lineage()

And let’s keep running the Cell Ranger pipeline in the background.
Show code cell content
transform = ln.Transform(
name="Preprocess Cell Ranger outputs", version="2.0", type="pipeline"
)
ln.track(transform)
[f.stage() for f in output_files]
filepath = ln.dev.datasets.schmidt22_perturbseq(basedir=ln.settings.storage)
file = ln.File(filepath, description="perturbseq counts")
file.save()
✅ saved: Transform(id='AcGXPfCKv0pRbb', name='Preprocess Cell Ranger outputs', version='2.0', type='pipeline', updated_at=2023-08-28 18:28:00, created_by_id='bKeW4T6E')
✅ saved: Run(id='CcwewUVHIwRRCLgERSdY', run_at=2023-08-28 18:28:00, transform_id='AcGXPfCKv0pRbb', created_by_id='bKeW4T6E')
💡 adding file g7iYzIUri8qUnH1gWnCb as input for run CcwewUVHIwRRCLgERSdY, adding parent transform JXkdP1eEWX0T9R
💡 adding file vSKTfjcMvRjK3s4ap0k1 as input for run CcwewUVHIwRRCLgERSdY, adding parent transform JXkdP1eEWX0T9R
💡 adding file OgvKbN8gXUfplSnstlMO as input for run CcwewUVHIwRRCLgERSdY, adding parent transform JXkdP1eEWX0T9R
💡 file in storage 'mydata' with key 'schmidt22_perturbseq.h5ad'
💡 data is AnnDataLike, consider using .from_anndata() to link var_names and obs.columns as features
Track app upload & analytics#
The hidden cell below simulates additional analytic steps including:
uploading phenotypic screen data
scRNA-seq analysis
analyses of the integrated datasets
Show code cell content
# app upload
ln.setup.login("testuser1")
transform = ln.Transform(name="Upload GWS CRISPRa result", type="app")
ln.track(transform)
filepath = ln.dev.datasets.schmidt22_crispra_gws_IFNG(ln.settings.storage)
file = ln.File(filepath, description="Raw data of schmidt22 crispra GWS")
file.save()
# upload and analyze the GWS data
ln.setup.login("testuser2")
transform = ln.Transform(name="GWS CRIPSRa analysis", type="notebook")
ln.track(transform)
file_wgs = ln.File.filter(key="schmidt22-crispra-gws-IFNG.csv").one()
df = file_wgs.load().set_index("id")
hits_df = df[df["pos|fdr"] < 0.01].copy()
file_hits = ln.File(hits_df, description="hits from schmidt22 crispra GWS")
file_hits.save()
✅ logged in with email testuser1@lamin.ai and id DzTjkKse
✅ saved: Transform(id='o0s9dpKHxvP14n', name='Upload GWS CRISPRa result', type='app', updated_at=2023-08-28 18:28:03, created_by_id='DzTjkKse')
✅ saved: Run(id='1Xv2nS3IhOJPAM0ufjHx', run_at=2023-08-28 18:28:03, transform_id='o0s9dpKHxvP14n', created_by_id='DzTjkKse')
💡 file in storage 'mydata' with key 'schmidt22-crispra-gws-IFNG.csv'
✅ logged in with email testuser2@lamin.ai and id bKeW4T6E
✅ saved: Transform(id='sZ0jO6lAxrrgfa', name='GWS CRIPSRa analysis', type='notebook', updated_at=2023-08-28 18:28:05, created_by_id='bKeW4T6E')
✅ saved: Run(id='7ZY75b0sn8jLMo36S2oP', run_at=2023-08-28 18:28:05, transform_id='sZ0jO6lAxrrgfa', created_by_id='bKeW4T6E')
💡 adding file UfHSSYBWcvwIuYcoP6JD as input for run 7ZY75b0sn8jLMo36S2oP, adding parent transform o0s9dpKHxvP14n
💡 file will be copied to default storage upon `save()` with key `None` ('.lamindb/4CXgRN4MkUHFT2hQzBHi.parquet')
💡 data is a dataframe, consider using .from_df() to link column names as features
✅ storing file '4CXgRN4MkUHFT2hQzBHi' at '.lamindb/4CXgRN4MkUHFT2hQzBHi.parquet'
Let’s see what the data lineage of this looks:
file = ln.File.filter(description="hits from schmidt22 crispra GWS").one()
file.view_lineage()

In the backgound, somebody integrated and analyzed the outputs of the app upload and the Cell Ranger pipeline:
Show code cell content
# Let us add analytics on top of the cell ranger pipeline and the phenotypic screening
transform = ln.Transform(
name="Perform single cell analysis, integrating with CRISPRa screen",
type="notebook",
)
ln.track(transform)
file_ps = ln.File.filter(description__icontains="perturbseq").one()
adata = file_ps.load()
screen_hits = file_hits.load()
import scanpy as sc
sc.tl.score_genes(adata, adata.var_names.intersection(screen_hits.index).tolist())
filesuffix = "_fig1_score-wgs-hits.png"
sc.pl.umap(adata, color="score", show=False, save=filesuffix)
filepath = f"figures/umap{filesuffix}"
file = ln.File(filepath, key=filepath)
file.save()
filesuffix = "fig2_score-wgs-hits-per-cluster.png"
sc.pl.matrixplot(
adata, groupby="cluster_name", var_names=["score"], show=False, save=filesuffix
)
filepath = f"figures/matrixplot_{filesuffix}"
file = ln.File(filepath, key=filepath)
file.save()
✅ saved: Transform(id='68JJSlWzF0zi5h', name='Perform single cell analysis, integrating with CRISPRa screen', type='notebook', updated_at=2023-08-28 18:28:05, created_by_id='bKeW4T6E')
✅ saved: Run(id='BSZSq23Y2fMYljD3TtTo', run_at=2023-08-28 18:28:05, transform_id='68JJSlWzF0zi5h', created_by_id='bKeW4T6E')
💡 adding file nj6Sg7LVgZyrA2RB7l0K as input for run BSZSq23Y2fMYljD3TtTo, adding parent transform AcGXPfCKv0pRbb
💡 adding file 4CXgRN4MkUHFT2hQzBHi as input for run BSZSq23Y2fMYljD3TtTo, adding parent transform sZ0jO6lAxrrgfa
WARNING: saving figure to file figures/umap_fig1_score-wgs-hits.png
💡 file will be copied to default storage upon `save()` with key 'figures/umap_fig1_score-wgs-hits.png'
✅ storing file '22s3gj55XIKJUj9IE0NR' at 'figures/umap_fig1_score-wgs-hits.png'
WARNING: saving figure to file figures/matrixplot_fig2_score-wgs-hits-per-cluster.png
💡 file will be copied to default storage upon `save()` with key 'figures/matrixplot_fig2_score-wgs-hits-per-cluster.png'
✅ storing file 'nEnEGke53hs6Cynn7yBw' at 'figures/matrixplot_fig2_score-wgs-hits-per-cluster.png'
The outcome of it are a few figures stored as image files. Let’s query one of them and look at the data lineage:
Track notebooks#
We’d now like to track the current Jupyter notebook to continue the work:
ln.track()
💡 notebook imports: ipython==8.14.0 lamindb==0.51.0 scanpy==1.9.4
✅ saved: Transform(id='1LCd8kco9lZUz8', name='Bird's eye view', short_name='birds-eye', version='0', type=notebook, updated_at=2023-08-28 18:28:07, created_by_id='bKeW4T6E')
✅ saved: Run(id='WZYNFjoMvznI2RFVMXCc', run_at=2023-08-28 18:28:07, transform_id='1LCd8kco9lZUz8', created_by_id='bKeW4T6E')
Visualize data lineage#
Let’s load one of the plots:
file = ln.File.filter(key__contains="figures/matrixplot").one()
from IPython.display import Image, display
file.stage()
display(Image(filename=file.path))
💡 adding file nEnEGke53hs6Cynn7yBw as input for run WZYNFjoMvznI2RFVMXCc, adding parent transform 68JJSlWzF0zi5h
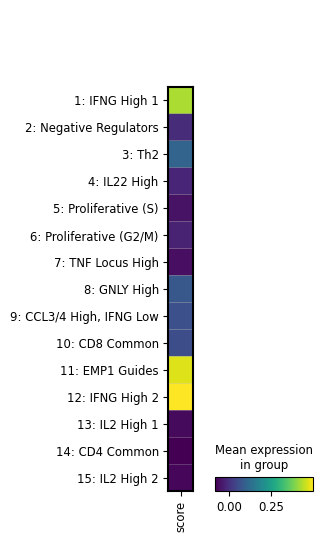
We see that the image file is tracked as an input of the current notebook. The input is highlighted, the notebook follows at the bottom:
file.view_lineage()

Alternatively, we can also purely look at the sequence of transforms and ignore the files:
transform = ln.Transform.search("Bird's eye view", return_queryset=True).first()
transform.parents.df()
| name | short_name | version | initial_version_id | type | reference | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|
| id | ||||||||
| 68JJSlWzF0zi5h | Perform single cell analysis, integrating with... | None | None | None | notebook | None | 2023-08-28 18:28:07 | bKeW4T6E |
transform.view_parents()

Understand runs#
We tracked pipeline and notebook runs through run_context, which stores a Transform and a Run record as a global context.
File objects are the inputs and outputs of runs.
What if I don’t want a global context?
Sometimes, we don’t want to create a global run context but manually pass a run when creating a file:
run = ln.Run(transform=transform)
ln.File(filepath, run=run)
When does a file appear as a run input?
When accessing a file via stage(), load() or backed(), two things happen:
The current run gets added to
file.input_ofThe transform of that file gets added as a parent of the current transform
You can then switch off auto-tracking of run inputs if you set ln.settings.track_run_inputs = False: Can I disable tracking run inputs?
You can also track run inputs on a case by case basis via is_run_input=True, e.g., here:
file.load(is_run_input=True)
Query by provenance#
We can query or search for the notebook that created the file:
transform = ln.Transform.search("GWS CRIPSRa analysis", return_queryset=True).first()
And then find all the files created by that notebook:
ln.File.filter(transform=transform).df()
| storage_id | key | suffix | accessor | description | version | initial_version_id | size | hash | hash_type | transform_id | run_id | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||
| 4CXgRN4MkUHFT2hQzBHi | lV7yGE5C | None | .parquet | DataFrame | hits from schmidt22 crispra GWS | None | None | 18368 | O2Owo0_QlM9JBS2zAZD4Lw | md5 | sZ0jO6lAxrrgfa | 7ZY75b0sn8jLMo36S2oP | 2023-08-28 18:28:05 | bKeW4T6E |
Which transform ingested a given file?
file = ln.File.filter().first()
file.transform
Transform(id='esPe9ey7UOWkUs', name='Chromium 10x upload', type='pipeline', updated_at=2023-08-28 18:27:59, created_by_id='DzTjkKse')
And which user?
file.created_by
User(id='DzTjkKse', handle='testuser1', email='testuser1@lamin.ai', name='Test User1', updated_at=2023-08-28 18:28:03)
Which transforms were created by a given user?
users = ln.User.lookup()
ln.Transform.filter(created_by=users.testuser2).df()
| name | short_name | version | initial_version_id | type | reference | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|
| id | ||||||||
| JXkdP1eEWX0T9R | Cell Ranger | None | 7.2.0 | None | pipeline | None | 2023-08-28 18:28:00 | bKeW4T6E |
| AcGXPfCKv0pRbb | Preprocess Cell Ranger outputs | None | 2.0 | None | pipeline | None | 2023-08-28 18:28:02 | bKeW4T6E |
| sZ0jO6lAxrrgfa | GWS CRIPSRa analysis | None | None | None | notebook | None | 2023-08-28 18:28:05 | bKeW4T6E |
| 68JJSlWzF0zi5h | Perform single cell analysis, integrating with... | None | None | None | notebook | None | 2023-08-28 18:28:07 | bKeW4T6E |
| 1LCd8kco9lZUz8 | Bird's eye view | birds-eye | 0 | None | notebook | None | 2023-08-28 18:28:07 | bKeW4T6E |
Which notebooks were created by a given user?
ln.Transform.filter(created_by=users.testuser2, type="notebook").df()
| name | short_name | version | initial_version_id | type | reference | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|
| id | ||||||||
| sZ0jO6lAxrrgfa | GWS CRIPSRa analysis | None | None | None | notebook | None | 2023-08-28 18:28:05 | bKeW4T6E |
| 68JJSlWzF0zi5h | Perform single cell analysis, integrating with... | None | None | None | notebook | None | 2023-08-28 18:28:07 | bKeW4T6E |
| 1LCd8kco9lZUz8 | Bird's eye view | birds-eye | 0 | None | notebook | None | 2023-08-28 18:28:07 | bKeW4T6E |
We can also view all recent additions to the entire database:
ln.view()
Show code cell output
File
| storage_id | key | suffix | accessor | description | version | initial_version_id | size | hash | hash_type | transform_id | run_id | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||
| nEnEGke53hs6Cynn7yBw | lV7yGE5C | figures/matrixplot_fig2_score-wgs-hits-per-clu... | .png | None | None | None | None | 28814 | JYIPcat0YWYVCX3RVd3mww | md5 | 68JJSlWzF0zi5h | BSZSq23Y2fMYljD3TtTo | 2023-08-28 18:28:07 | bKeW4T6E |
| 22s3gj55XIKJUj9IE0NR | lV7yGE5C | figures/umap_fig1_score-wgs-hits.png | .png | None | None | None | None | 118999 | laQjVk4gh70YFzaUyzbUNg | md5 | 68JJSlWzF0zi5h | BSZSq23Y2fMYljD3TtTo | 2023-08-28 18:28:07 | bKeW4T6E |
| 4CXgRN4MkUHFT2hQzBHi | lV7yGE5C | None | .parquet | DataFrame | hits from schmidt22 crispra GWS | None | None | 18368 | O2Owo0_QlM9JBS2zAZD4Lw | md5 | sZ0jO6lAxrrgfa | 7ZY75b0sn8jLMo36S2oP | 2023-08-28 18:28:05 | bKeW4T6E |
| UfHSSYBWcvwIuYcoP6JD | lV7yGE5C | schmidt22-crispra-gws-IFNG.csv | .csv | None | Raw data of schmidt22 crispra GWS | None | None | 1729685 | cUSH0oQ2w-WccO8_ViKRAQ | md5 | o0s9dpKHxvP14n | 1Xv2nS3IhOJPAM0ufjHx | 2023-08-28 18:28:04 | DzTjkKse |
| nj6Sg7LVgZyrA2RB7l0K | lV7yGE5C | schmidt22_perturbseq.h5ad | .h5ad | AnnData | perturbseq counts | None | None | 20659936 | la7EvqEUMDlug9-rpw-udA | md5 | AcGXPfCKv0pRbb | CcwewUVHIwRRCLgERSdY | 2023-08-28 18:28:02 | bKeW4T6E |
| vSKTfjcMvRjK3s4ap0k1 | lV7yGE5C | perturbseq/filtered_feature_bc_matrix/features... | .tsv.gz | None | None | None | None | 6 | kp6xn-SIwkaFI-xDC7Njhw | md5 | JXkdP1eEWX0T9R | gXXVcK1otXqE6UHukFqd | 2023-08-28 18:28:00 | bKeW4T6E |
| OgvKbN8gXUfplSnstlMO | lV7yGE5C | perturbseq/filtered_feature_bc_matrix/matrix.m... | .mtx.gz | None | None | None | None | 6 | bAkxpeZNXSoBfxLt1VXeZA | md5 | JXkdP1eEWX0T9R | gXXVcK1otXqE6UHukFqd | 2023-08-28 18:28:00 | bKeW4T6E |
Run
| transform_id | run_at | created_by_id | reference | reference_type | |
|---|---|---|---|---|---|
| id | |||||
| vYYOHgAJkgeRJIuStm1D | esPe9ey7UOWkUs | 2023-08-28 18:27:59 | DzTjkKse | None | None |
| gXXVcK1otXqE6UHukFqd | JXkdP1eEWX0T9R | 2023-08-28 18:28:00 | bKeW4T6E | None | None |
| CcwewUVHIwRRCLgERSdY | AcGXPfCKv0pRbb | 2023-08-28 18:28:00 | bKeW4T6E | None | None |
| 1Xv2nS3IhOJPAM0ufjHx | o0s9dpKHxvP14n | 2023-08-28 18:28:03 | DzTjkKse | None | None |
| 7ZY75b0sn8jLMo36S2oP | sZ0jO6lAxrrgfa | 2023-08-28 18:28:05 | bKeW4T6E | None | None |
| BSZSq23Y2fMYljD3TtTo | 68JJSlWzF0zi5h | 2023-08-28 18:28:05 | bKeW4T6E | None | None |
| WZYNFjoMvznI2RFVMXCc | 1LCd8kco9lZUz8 | 2023-08-28 18:28:07 | bKeW4T6E | None | None |
Storage
| root | type | region | updated_at | created_by_id | |
|---|---|---|---|---|---|
| id | |||||
| lV7yGE5C | /home/runner/work/lamin-usecases/lamin-usecase... | local | None | 2023-08-28 18:27:58 | DzTjkKse |
Transform
| name | short_name | version | initial_version_id | type | reference | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|
| id | ||||||||
| 1LCd8kco9lZUz8 | Bird's eye view | birds-eye | 0 | None | notebook | None | 2023-08-28 18:28:07 | bKeW4T6E |
| 68JJSlWzF0zi5h | Perform single cell analysis, integrating with... | None | None | None | notebook | None | 2023-08-28 18:28:07 | bKeW4T6E |
| sZ0jO6lAxrrgfa | GWS CRIPSRa analysis | None | None | None | notebook | None | 2023-08-28 18:28:05 | bKeW4T6E |
| o0s9dpKHxvP14n | Upload GWS CRISPRa result | None | None | None | app | None | 2023-08-28 18:28:04 | DzTjkKse |
| AcGXPfCKv0pRbb | Preprocess Cell Ranger outputs | None | 2.0 | None | pipeline | None | 2023-08-28 18:28:02 | bKeW4T6E |
| JXkdP1eEWX0T9R | Cell Ranger | None | 7.2.0 | None | pipeline | None | 2023-08-28 18:28:00 | bKeW4T6E |
| esPe9ey7UOWkUs | Chromium 10x upload | None | None | None | pipeline | None | 2023-08-28 18:27:59 | DzTjkKse |
User
| handle | name | updated_at | ||
|---|---|---|---|---|
| id | ||||
| bKeW4T6E | testuser2 | testuser2@lamin.ai | Test User2 | 2023-08-28 18:28:05 |
| DzTjkKse | testuser1 | testuser1@lamin.ai | Test User1 | 2023-08-28 18:28:03 |
Show code cell content
!lamin login testuser1
!lamin delete --force mydata
!rm -r ./mydata
✅ logged in with email testuser1@lamin.ai and id DzTjkKse
💡 deleting instance testuser1/mydata
✅ deleted instance settings file: /home/runner/.lamin/instance--testuser1--mydata.env
✅ instance cache deleted
✅ deleted '.lndb' sqlite file
❗ consider manually deleting your stored data: /home/runner/work/lamin-usecases/lamin-usecases/docs/mydata
